Project Leadership
Ben Bowen

Application Lead: In the Environmental Genomics and Systems Biology (EGSB) Division of Berkeley Lab, Ben is the Applications Lead for OpenMSI. He has over 10 years experience in mass spectrometry and quantitative analysis. With a background in chemical engineering and a biophysics PhD in multiparameter imaging of single-molecule fluorescence he has gone on to establish new paradigms in proteomics, metabolomics, and imaging.
Leading OpenMSI is a long-standing vision of Ben's where the power of web-based technologies allows the most advanced analysis, management, and visualization of one of the most promising technical approaches emerging into the biological laboratories.
Oliver Rübel

Computing Lead: Oliver is a research scientist of Berkeley Lab's Data Analytics and Visualization Group and a member of the NERSC Analytics team. He received his Ph.D. in Computer Science in 2009 from the University of Kaiserslautern, Germany. His research has focused on high-performance data analysis and visualization, machine learning, and query-driven visualization of multi-dimensional scientific data. During his career, Oliver has worked closely with applications ranging from high-energy physics, climate science, to biological sciences. He has contributed to the development of many visualization and analysis codes and has been a main developer of PointCloudXplore . With OpenMSI he continues his journey to make complex data accessible to application sciences. For more information about Oliver see here
Development Team
Curt Fischer

Senior Developer: Curt studied chemical engineering as an undergrad at Case Western Reserve University and as a Ph.D. student at MIT. His doctoral thesis, explored experimental approaches to optimizing metabolic pathways for microbial hydrocarbon and biofuel production. From 2009 to 2010 he was a post-doctoral scholar at UC Berkeley, where he studied natural microbial communities using a combination of -omics approaches. From 2011-2014, he worked as a Biological Engineer at startup company Ginkgo Bioworks, where he led projects enabling bacteria to convert CO2 to fuels, helped expand the company's organism testing and characterization pipeline, and translated client needs into biological specifications. In 2015 he joined the OpenMSI team.
Annette M. Greiner

Senior Developer: Annette Greiner is a user interface designer and web developer with particular interest in interaction design and visualization. She recently earned a Master's in Information Management and Systems from UC Berkeley's School of Information, with a focus on human-computer interaction. Before returning to school, she developed web sites for the Advanced Light Source and the DOE Joint Genome Institute. At NERSC, she designs and develops science gateways. Annette holds a B.S. in biomedical science and theater from the University of Michigan.
Shreyas Cholia

Senior Developer: Shreyas Cholia works on science gateway, web and grid technologies for NERSC, with the goal of making high-performance and distributed computing more transparent and accessible. He has also been involved in various grid and data-driven science efforts at NERSC since 2002. Prior to his appointment at NERSC, Shreyas was a developer and consultant at IBM. He has a bachelor's degree from Rice University, where he double majored in Computer Science and Cognitive Sciences.
OpenMSI Advisory Board
Katherine Yelick

Katherine Yelick is a Professor of Electrical Engineering and Computer Sciences at the University of California at Berkeley and is also the Associate Laboratory Director of Computing Sciences at Lawrence Berkeley National Laboratory.
Gary Karpen

Gary Karpen is an Adjunct Professor with the Department of Molecular and Cell Biology at the University of California at Berkeley and is also the Division Director of the Life Sciences Division at Lawrence Berkeley National Laboratory
Trent Northen

Trent Northen is a Staff Scientist at Lawrence Berkeley National Laboratory (LBNL) and lead of the Molecular Cartography Project at LBNL.
David Skinner

David Skinner is Group Lead of the Outreach, Software and Programming Group at the National Energy Scientific Computing Center (NERSC).
Wes Bethel

Wes Bethel is the Group Lead of the Visualization Group at Lawrence Berkeley National Laboratory.
Ken Joy

Ken Joy is a Professor in the Computer Science Department at the University of California at Davis. He is the Director of the Institute for Data Analysis and Visualization (IDAV) at UC Davis.
Overview
Specific Goal: To make the most high-performance, advanced data management, model building, analysis and visualization resources for mass spectrometry imaging accessible to scientists via the web.
Project summary: The development and application of cutting-edge analytical methods is a core driver for new scientific discoveries, medical diagnostics, and commercial-innovation. Mass spectrometry imaging (MSI) holds the promise for being a transformative technology for advanced studies of metabolic processes with broad applications in life sciences, bioenergy, and health. MSI enables fast assays of microbial metabolism essential in bioengineering problems common to the development of biofuels, drugs, and diagnostics for cancer and other diseases. While the data can be routinely collected, the broad application of MSI is currently limited by the lack of easily accessible analysis methods that can process data of the size, volume, diversity and complexity generated by MSI experiments. Typically individuals that can both develop quality computer software and also understand the biological and technical issues associated with each experiment are required to help even with common, and what ought to be routine analyses. Furthermore, the expert developer is often handicapped due to a lack of standards in data-format, algorithms, and visualization strategies. This is an unfortunate situation in which well-designed experiments have to wait in the queue (often for weeks or month) until someone is available to help with the analysis, in which experiments are not completed due to delayed detection of errors, and in which advanced, large-scale experiments are not performed due to the lack of essential analysis capabilities. Analysis of MSI data is an unsolved big data problem that slows down and prevents high-impact scientific discoveries.
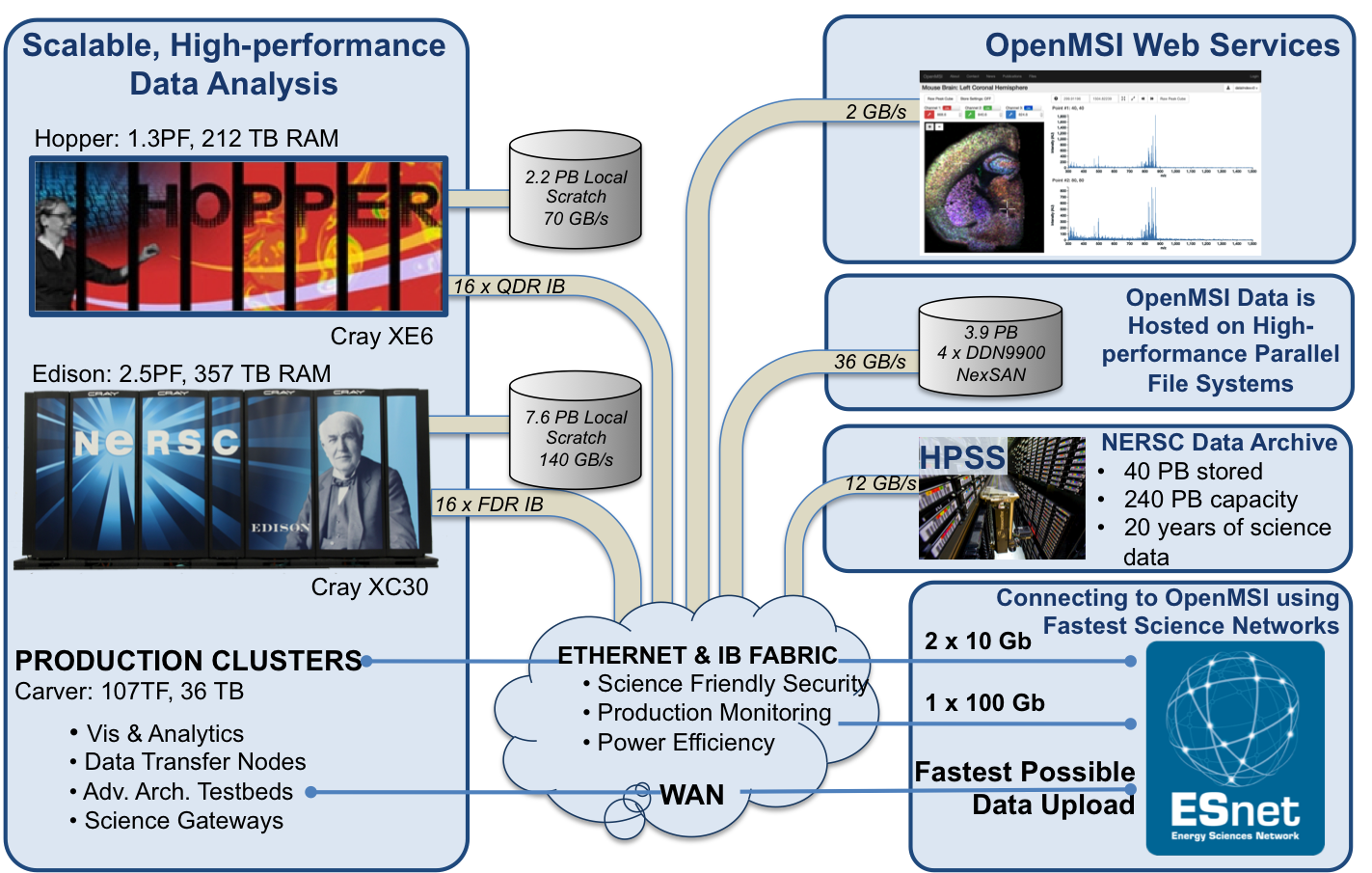 Advanced, high-performance computing: OpenMSI uses the advanced, high-performance compute resources of the National Energy Research Scientific Computing Center (NERSC) to make advanced mass spectrometry imaging accessible. The combination of advanced software solutions and state-of-the-art computing are what makes OpenMSI possible.
Advanced, high-performance computing: OpenMSI uses the advanced, high-performance compute resources of the National Energy Research Scientific Computing Center (NERSC) to make advanced mass spectrometry imaging accessible. The combination of advanced software solutions and state-of-the-art computing are what makes OpenMSI possible.
Citing OpenMSI
Oliver Rübel, Annette Greiner, Shreyas Cholia, Katherine Louie, E. Wes Bethel, Trent R. Northen, and Benjamin P. Bowen, "OpenMSI: A High-Performance Web-Based Platform for Mass Spectrometry Imaging" Analytical Chemistry 2013 85 (21), 10354-10361, DOI: 10.1021/ac402540a. [BibTeX][Online at ACS]