OpenMSI: Web-based visualization, analysis and management of mass spectrometry imaging data
Mass spectrometry imaging (MSI) is widely applied to image complex samples for applications spanning health, microbial ecology, and high throughput screening of high-density arrays. MSI has emerged as a technique suited to resolving metabolism within complex cellular systems; where understanding the spatial variation of metabolism is vital for making a transformative impact on science. Unfortunately, the scale of MSI data and complexity of analysis presents an insurmountable barrier to scientists where a single 2D-image may be many gigabytes and comparison of multiple images is beyond the capabilities available to most scientists. The OpenMSI project will overcome these challenges, allowing broad use of MSI to researchers by providing a web-based gateway for management and storage of MSI data, the visualization of the hyper-dimensional contents of the data, and the statistical analysis.
Select an image below or click here to view additional files
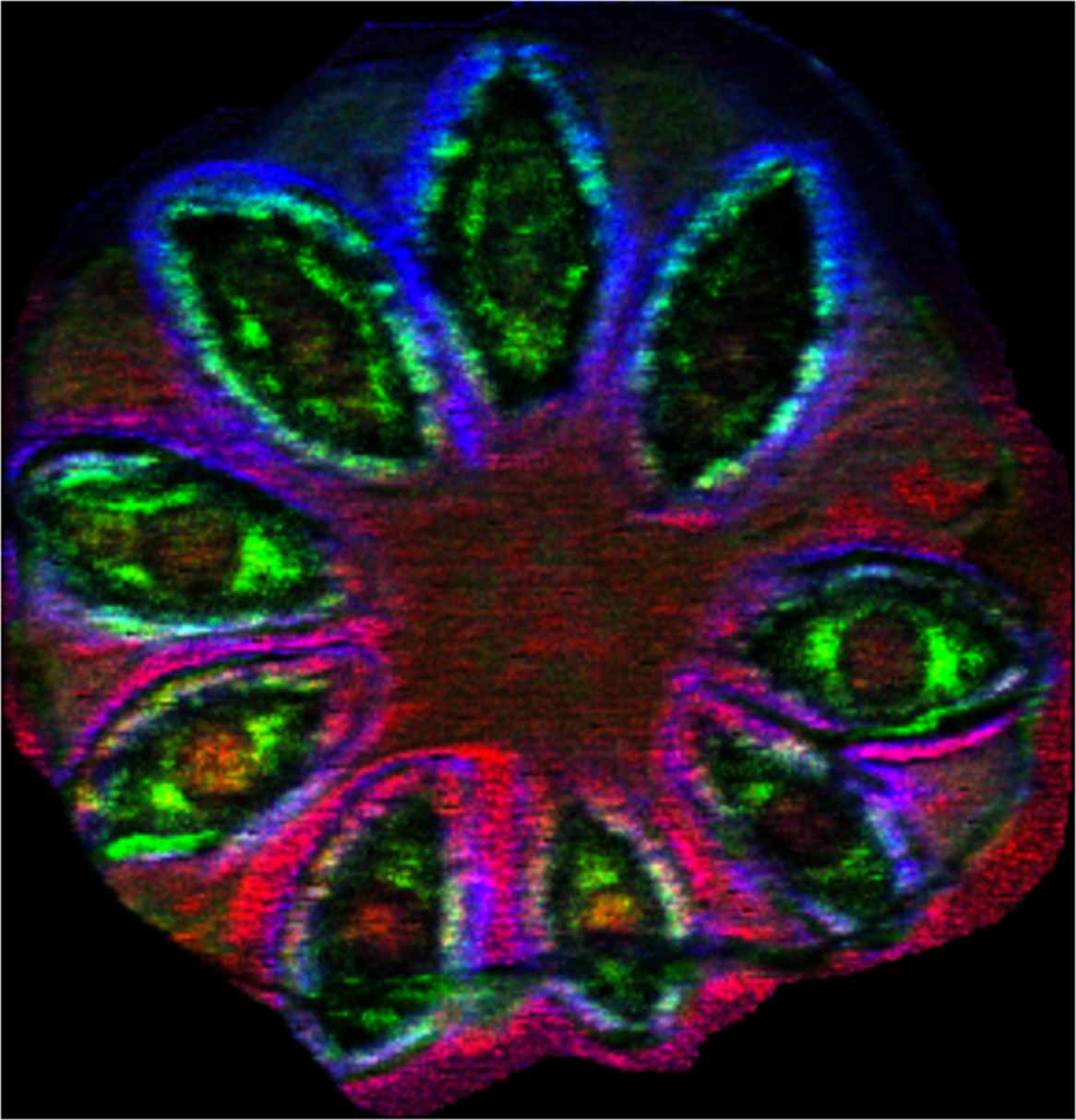
Sample: Flax (Linum usitatissimum) pod
Instrument: Waters Synapt G2 HDMS
Desorption/Ionization Method: MALDI DHB
Original File: *.imzML and *.img
Original Filesize: 15 GB
OpenMSI Filesize: 2.7 GB
Image dimensions: 303 x 291 pixels
Pixel size: 20 microns
#m/z bins: 90,000
Contributors: Doralyn S. Dalisay, Hong Yang, Laurence B. Davin and Norman G. Lewis
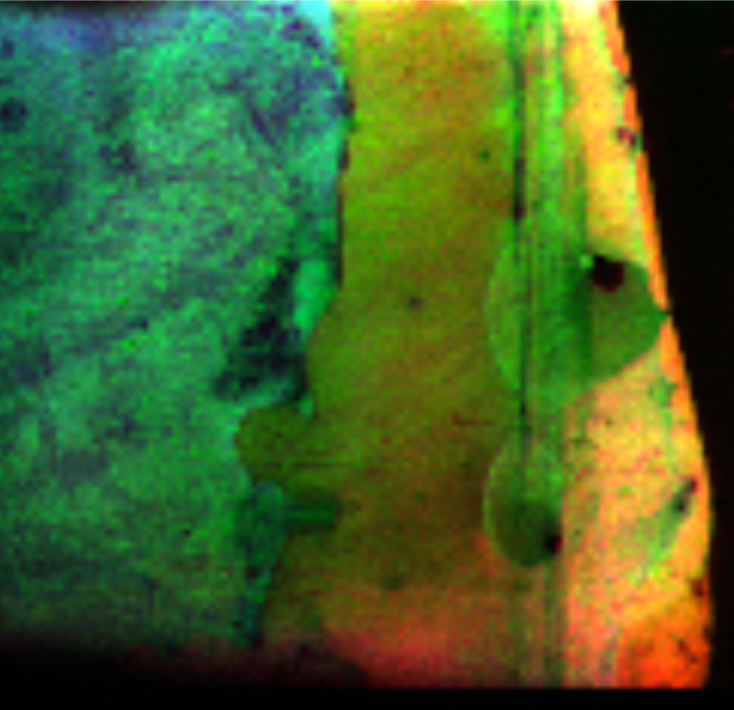
Sample: Potato eye stamped on NIMS chip
Instrument: AbSciex 5800
Desorption/Ionization Method: NIMS
Original File: *.img
Original Filesize: 21 GB
OpenMSI Filesize: 8.5 GB
Image dimensions: 123 x 463 pixels
Pixel size: 100 microns
#m/z bins: 188,960
Computed data: peak-finding, NMF
Contributors: Katherine Louie and Trent Northen
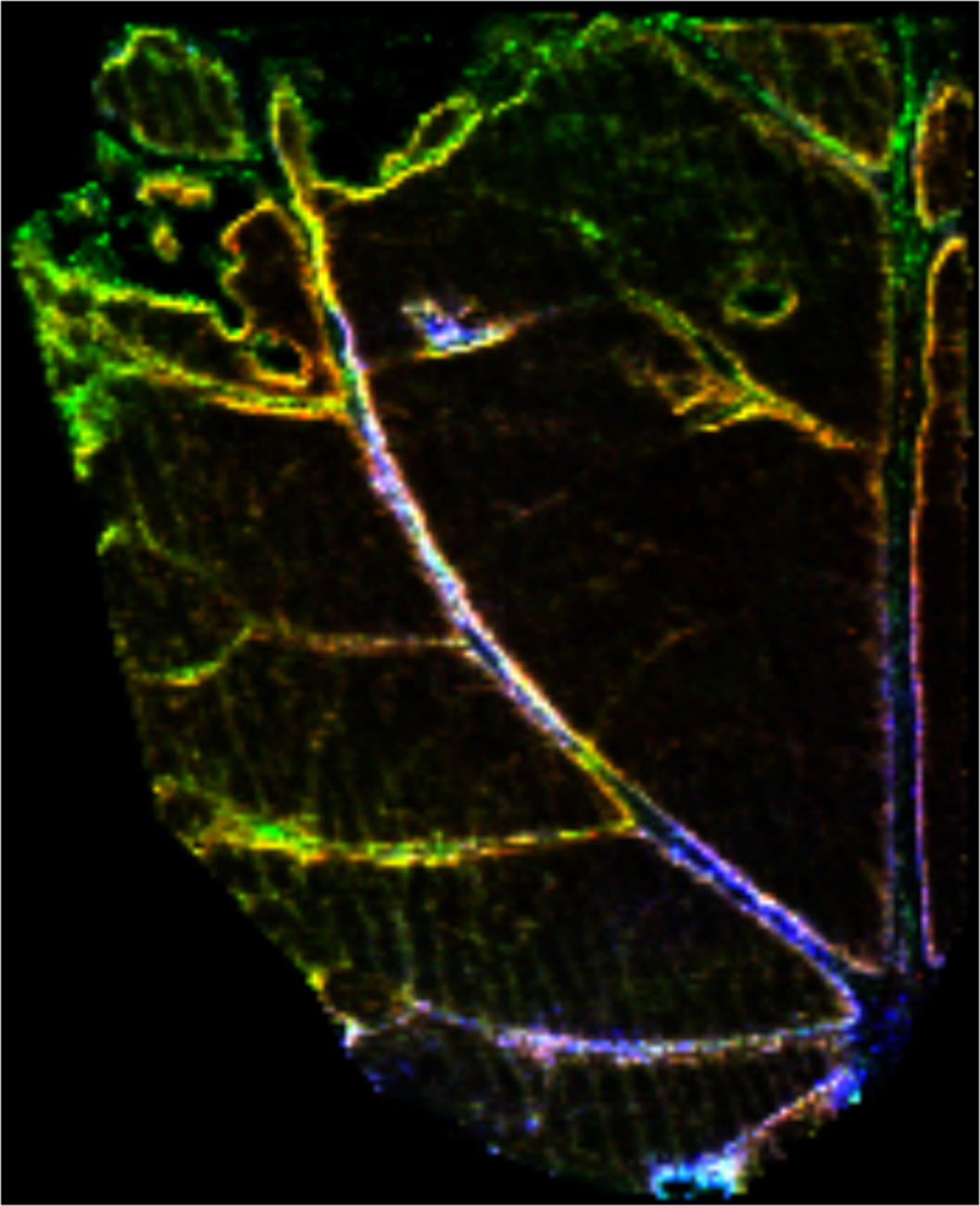
Sample: Leaf of transgenic hybird Poplar (Populus tremula × Populus alba)
Instrument: Waters Synapt G2 HDMS
Desorption/Ionization Method: MALDI DHB
Original File: *.img and *.imzML
Original Filesize: 4.2 GB
OpenMSI Filesize: 543 MB
Image dimensions: 213 x 173 pixels
Pixel size: 50 microns
#m/z bins: 60,000
Contributors: Costa, M.A., Marques, J.V., Dalisay, D.S., Herman, B., Bedgar, D.L., Davin, L.B. and Lewis N.G. Transgenic hybrid poplar for sustainable and scalable production of the commodity/specialty chemical, 2-phenylethanol. PLoS One, 2013 Dec 26;8(12):e83169

Sample: two microbes plated on rich LB-agar
Instrument: AbSciex 5800
Desorption/Ionization Method: NIMS
Original File: *.img
Original Filesize: 8.8 GB
OpenMSI Filesize: 1.2 GB
Image dimensions: 250 x 160 pixels
Pixel size: 100 microns
#m/z bins: 116,152
Contributors: Louie, K. B.; Bowen, B. P.; Cheng, X.; Berleman, J. E.; Chakraborty, R.; Deutschbauer, A.; Arkin, A.; Northen, T. R. “Replica-extraction-transfer” nanostructure-initiator mass spectrometry imaging of acoustically printed bacteria. Anal. Chem. 2013, 85, 10856–10862.
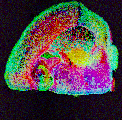
Sample: a mouse brain: left coronal hemisphere
Instrument: AbSciex 5800
Desorption/Ionization Method: NIMS
Original File: *.img
Original Filesize: 2.2 GB
OpenMSI Filesize: 0.3 GB
Image dimensions: 122 x 120 pixels
Pixel size: 80 microns
#m/z bins: 80,339
Computed data: peak-finding, NMF
Contributors: Katherine Louie, Michael Balamotis, Ben Bowen, and Trent Northen
Sample: a rat lung
Instrument: Thermo LTQ-Orbitrap
Desorption/Ionization Method: MALDI
Original File: *.RAW
Original Filesize: 1 GB
OpenMSI Filesize: 660 MB
Image dimensions: 132 x 149 pixels
Pixel size: 50 microns
#m/z bins: 500,000
Contributors: Fehniger, T. E.; Végvári, Á.; Rezeli, M.; Prikk, K.; Ross, P.; Dahlbäck, M.; Edula, G.; Sepper, R.; Marko-Varga, G. Direct Demonstration of Tissue Uptake of an Inhaled Drug: Proof-of-Principle Study Using Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging. Anal. Chem. 2011, 83, 8329–8336.